Ecological Nitrogen Limitation Shapes the DNA Composition of Plant Genomes
Molecular Biology and Evolution | 26:953–956
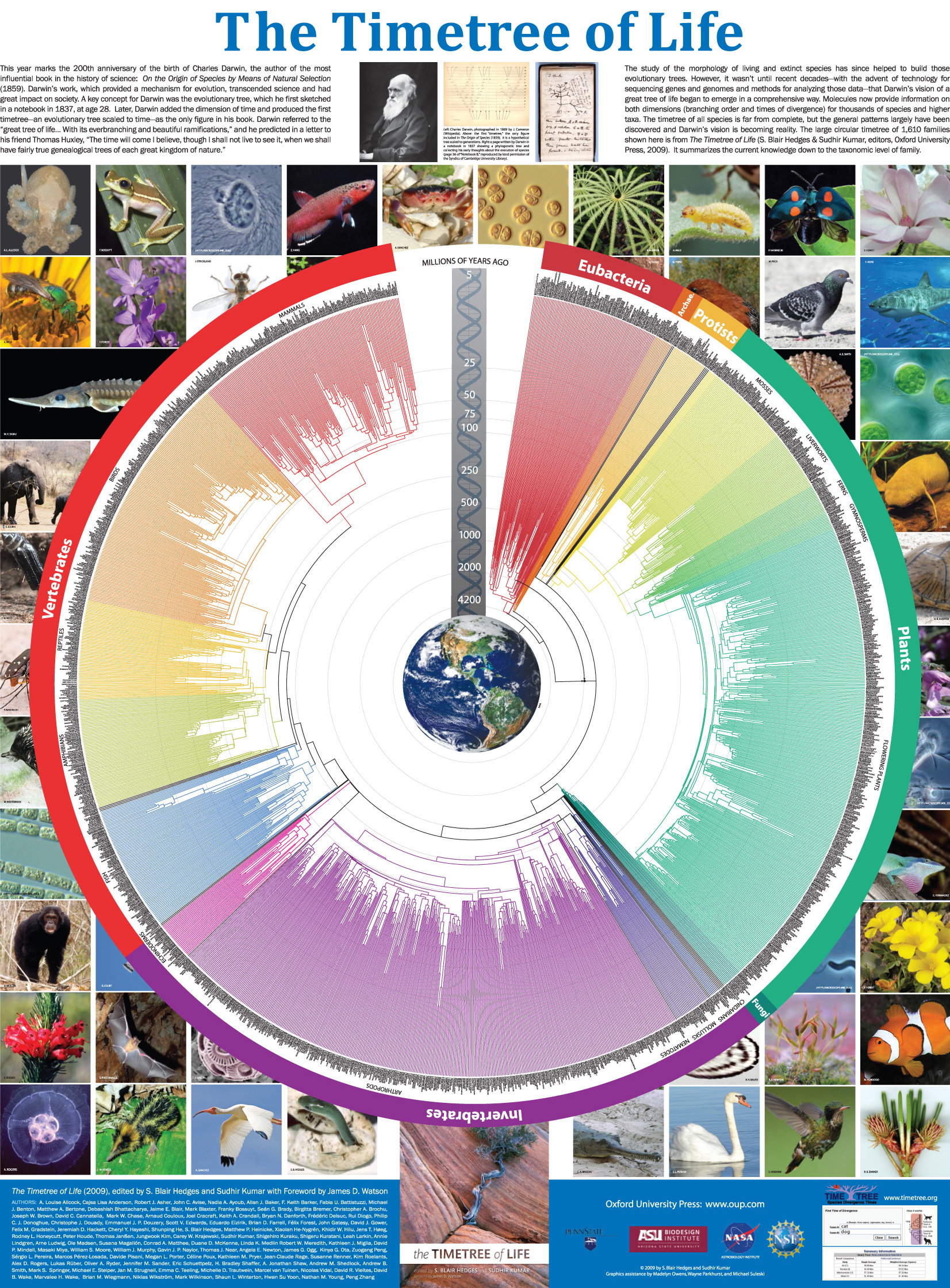
The Timetree of Life
Oxford University Press (New York) | (551 pp)

Discovering the Timetree of Life
The TimeTree of Life (ed. Hedges, Kumar) [Oxford University Press] | 3–18
Signatures of Nitrogen Limitation in the Elemental Composition of the Proteins Involved in the Metabolic Apparatus
Proceedings of the Royal Society B: Biological Sciences | 276:2605–2610
Association of Orthodenticle with Natural Variation for Early Embryonic Patterning in Drosophila melanogaster
Journal of Experimental Zoology Part B: Molecular and Developmental Evolution | 312:841–854
Phylogenetic Construction of 17 Bacterial Phyla by New Method and Carefully Selected Orthologs
Gene | 429:59–64
A Bag-of-words Approach for Drosophila Gene Expression Pattern Annotation
BMC Bioinformatics | 10:119 (16 pp)
Drosophila Gene Expression Pattern Annotation Using Sparse Features and Term-term Interactions
KDD-09: 15th ACM SIGKDD Conference on Knowledge Discovery and Data Mining | 407–415
Positional Conservation and Amino Acids Shape the Correct Diagnosis and Population Frequencies of Benign and Damaging Personal Amino Acid Mutations
Genome Research | 19:1562–1569
Relationship between Gene Co-expression and Sharing of Transcription Factor Binding Sites in Drosophila Melanogasterc
Bioinformatics | 25:2473–2477
The Origin of Metazoa: A Transition from Temporal to Spatial Cell Differentiation
BioEssays | 31:758–768
Methods for Incorporating the Hypermutability of CpG Dinucleotides in Detecting Natural Selection Operating at the Amino Acid Sequence Level
Molecular Biology and Evolution | 26:2275–2284
Developmental Stage Annotation of Drosophila Gene Expression Pattern Images via an Entire Solution Path for LDA
ACM Transactions on Knowledge Discovery from Data | 2:(21 pp)
Biological Image Analysis via Matrix Approximation
Encyclopedia of Data Warehousing and Mining (ed. Wang) [Idea Group] | 166–170
Automated Annotation of Drosophila Gene Expression Patterns Using a Controlled Vocabulary
Bioinformatics | 24:1881–1888
MEGA: A Biologist-centric Software for Evolutionary Analysis of DNA and Protein Sequences
Briefings in Bioinformatics | 9:299–306
Detecting Molecular Signatures of Adaptive Evolution
Evolutionary Genomics and Proteomics (ed. Pagel, Pomiankowski) [Sinauer] | 241–254
Nullomers: Really a Matter of Natural Selection?
PLoS One | 2:1022–1025
Evolution of Genes and Genomes on the Drosophila Phylogeny
Nature | 450:203–218
Lower Bounds on Multiple Sequence Alignment Using Exact 3-way Alignment
BMC Bioinformatics | 8:140 (8 pp)
Bioinformatics Software for Biologists in the Genomics Era
Bioinformatics | 23:1713–1717
Multiple Sequence Alignment: In Pursuit of Homologous DNA Positions
Genome Research | 17:127–135
MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) Software Version 4.0
Molecular Biology and Evolution | 24:1596–1599
Constraint and Turnover in Sex-biased Gene Expression in the Genus Drosophila
Nature | 450:233–237
Classification of Drosophila Embryonic Developmental State Range Based on Gene Expression Pattern Images
Computational Systems Bioinformatics Conference | 2006:293–298
Codon-based Detection of Positive Selection Can be Biased by Heterogeneous Distribution of Polar Amino Acids along Protein Sequences
Computational Systems Bioinformatics Conference | 2006:335–340
Signatures of Ecological Resource Availability in the Animal and Plant Proteomes
Molecular Biology and Evolution | 23:1946–1951
TimeTree: A Public Knowledge-base of Divergence Times among Organisms
Bioinformatics | 22:2971–2972
Constraining Fossil Calibrations for Molecular Clocks
BioEssays | 28:770–771
Evolutionary Anatomies of Positions and Types of Disease-associated and Neutral Amino Acid Mutations in the Human Genome
BMC Genomics | 7:306 (9 pp)
Higher Intensity of Purifying Selection on >90% of the Human Genes Revealed by the Intrinsic Replacement Mutation Rates
Molecular Biology and Evolution | 23:2283–2287
The Evolution of the Genome: Comparative Genomics in Eukaryotes
The Evolution of the Genome (ed. Gregory) [Elsevier] | 521–583
Maximum Likelihood Outperforms Maximum Parsimony Even When Evolutionary Rates are Heterotachous
Molecular Biology and Evolution | 22:2139–2141
Inferring Species Phylogenies from Multiple Genes: Concatenated Sequence Tree versus Consensus Gene Tree
Journal of Experimental Zoology Part B: Molecular and Developmental Evolution | 304:64–74
Automatic Annotation Techniques for Gene Expression Images of the Fruit Fly Embryo
Visual Communications and Image Processing 2005, Pts 1-4 | 5960:576–583
Molecular Clocks: Four Decades of Evolution
Nature Reviews Genetics | 6:654–662
Placing Confidence Limits on the Molecular Age of the Human-Chimpanzee Divergence
Proceedings of the National Academy of Sciences (USA) | 102:18842–18847
Pushing Back the Expansion of Introns in Animal Genomes
Cell | 123:1182–1184
Image Registration and Similarity Computation for Chicken Gene Expression Patterns
Genomic Signal Processing and Statistics | (4 pp)
The Spectrum of Human Rhodopsin Disease Mutations through the Lens of Interspecific Variation
Gene | 12:107–118
Identifying Spatially Similar Gene Expression Patterns in Early Stage Fruit Fly Embryo Images: Binary Feature versus Invariant Moment Digital Representations
BMC Bioinformatics | 16:202–214
Precision of Molecular Time Estimates
Trends in Genetics | 20:242–247
Gene Expression Intensity Shapes Evolutionary Rates of the Proteins Encoded by the Vertebrate Genome
Genetics | 168:373–381
Prospects for Inferring Very Large Phylogenies by Using the Neighbor-joining Method
Proceedings of the National Academy of Sciences (USA) | 101:11030–11035
Temporal Patterns of Fruit Fly (Drosophila) Evolution Revealed by Mutation Clocks
Molecular Biology and Evolution | 21:36–44
MEGA3: Integrated Software for Molecular Evolutionary Genetics Analysis and Sequence Alignment
Briefings in Bioinformatics | 5:150–163
Genomic Clocks and Evolutionary Timescales
Trends in Genetics | 19:200–206
Genomic Sequence of a Ranavirus (family Iridoviridae) Associated with Salamander Mortalities in North America
Virology | 316:90–103
Quantifying the Intragenic Distribution of Human Disease Mutations
Annals of Human Genetics | 67:567–579
Heterogeneity of Nucleotide Frequencies among Evolutionary Lineages and Phylogenetic Inference
Molecular Biology and Evolution | 20:610–621
Taxon Sampling, Bioinformatics, and Phylogenomics
Systematic Biology | 52:119–124
Patterns of Transitional Mutation Biases within and among Mammalian Genomes
Molecular Biology and Evolution | 20:988–993
Neutral Substitutions Occur at a Faster Rate in Exons than in Noncoding DNA in Primate Genomes
Genome Research | 13:838–844
Vertebrate Genomes Compared
Science | 297:1283–1285
Measuring Conservation of Contiguous Sets of Autosomal Markers on Bovine and Porcine Genomes in Relation to the Map of the Human Genome
Genome | 45:769–776
BEST: A Novel Computational Approach for Comparing Gene Expression Patterns from Early Stages of Drosophila melanogaster Development
Genetics | 162:2037–2047
Mutation Rates in Mammalian Genomes
Proceedings of the National Academy of Sciences (USA) | 99:803–808
Evolutionary Distance Estimation under Heterogeneous Substitution Pattern among Lineages
Molecular Biology and Evolution | 19:1727–1736
Elucidating Gene Interaction Networks Based on Gene Expression Pattern Image Analysis
Proceedings of the International Conference on Biomedical Engineering | 5:232–234
Does Nonneutral Evolution Shape Observed Patterns of DNA Variation in Animal Mitochondrial Genomes?
Annual Review of Genetics | 35:539–566
A Genomic Timescale for the Origin of Eukaryotes
BMC Evolutionary Biology | 1:4 (10 pp)
Mutation and Linkage Disequilibrium in Human mtDNA
European Journal of Human Genetics | 9:969–972
Classification and Indexing of Gene Expression Images
Applications of Digital Image Processing XXIV | 4472:471–481
Disparity Index: A Simple Statistic to Measure and Test the Homogeneity of Substitution Patterns between Molecular Sequences
Genetics | 158:1321–1327
Determination of the Number of Conserved Chromosomal Segments between Species
Genetics | 157:1387–1395
MEGA2: Molecular Evolutionary Genetics Analysis Software
Bioinformatics | 17:1244–1245
Understanding Human Disease Mutations through the Use of Interspecific Genetic Variation
Human Molecular Genetics | 10:2319–2328
Incomplete Taxon Sampling is Not a Problem for Phylogenetic Inference
Proceedings of the National Academy of Sciences (USA) | 98:10751–10756
Traditional Phylogenetic Reconstruction Methods Reconstruct Shallow and Deep Evolutionary Relationships Equally Well
Molecular Biology and Evolution | 18:1823–1827
Efficiency of the Neighbor-joining Method in Reconstructing Deep and Shallow Evolutionary Relationships in Large Phylogenies
Journal of Molecular Evolution | 51:544–553
Questioning Evidence for Recombination in Human Mitochondrial DNA
Science | 288:1931a (2 pp)
Expansion and Molecular Evolution of the Interferon-induced 2 '-5 ' Oligoadenylate Synthetase Gene Family
Molecular Biology and Evolution | 17:738–750
Single Column Discrepancy and Dynamic Max-Mini Optimizations for Quickly Finding the Most Parsimonious Evolutionary Trees
Bioinformatics | 16:140–151