The Genetic Foundations of Convergent Traits
Nature Reviews Genetics | accepted
Entrenchment and Contingency in Neutral Protein Evolution with Epistasis
Genome Biology | in revision
Nutrigenomics: From Epicure to Epigenetics - Engaging Students with Nutrition Through the Human Genome
The American Biology Teacher | accepted
Treemble: A Graphical Tool to Generate Newick Strings from Phylogenetic Tree Images
Bioinformatics | in review
An Evolutionary Metric for Estimating PhyloAges from Bulk Sequencing of Hematopoietic Stem Cells Reveals the Tempo of Blood Aging in Cancer and Longevity
Journal of Molecular Evolution | (13 pp)
Evolutionary dynamics of early and late mutational signatures in metastatic cancer
Frontiers in Bioinformatics | in review
Transformer-based Hi-C Upscaling Reveals 3D Genome Structure at Low-resolution Sequencing
Cell Genomics | (submitted)
The Big Evolution of the Little Phage: Teaching biological interdependency through a virus
The American Biology Teacher) | in press
A collaborative genomics platform for T1D research in Eastern and Central Europe
Metabologia | in submission
R3F: An R Package for Evolutionary Dates, Rates, and Priors using Relative Rate Framework
arXiv | (available)
Enabling Data-driven Discoveries in Evolutionary Genetics and Genomics
Genetics | 230:iyaf084 (4 pp)
MEGA 12.1: Cross-platform release for macOS and Linux operating systems
Journal of Molecular Evolution | (5 pp)
Evolutionary Sparse Learning Reveals the Shared Genetic Basis of Convergent Traits
Nature Communications | 16:3217 (14 pp)
Robust and Efficient Method for Estimating Confidence Limits for Phylogenomic Inferences of Organismal Relationships
Molecular Biology and Evolution | 42:msaf296 (11 pp)
MEGA-GPT: Artificial Intelligence Guidance and Building Analytical Protocols using MEGA software
Molecular Biology and Evolution | 42:msaf101 (5 pp)
Genome Assembly of Arctica islandica, the Longest-Lived Non-Colonial Animal Species
Animals | 15:ani15050690 (13 pp)
MyESL: A software for Evolutionary Sparse Learning in Molecular Phylogenetics and Genomics
Molecular Biology and Evolution | 42:msaf224 (16 pp)
Integrating Phylogenies with Chronology to Assemble the Tree of Life
Frontiers in Bioinformatics | 5:1571568 (14 pp)
STICI: Split-Transformer with Integrated Convolutions for Genotype Imputation
Nature Communications | 16:(14 pp)
Teaching Biology & Evolution Through the Watery Matrix of Life: A Genomic Medicine Perspective
The American Biology Teacher | 87:84–92
Completing a Molecular Timetree of Afrotheria
Frontiers in Bioinformatics | 5:1710926 (9 pp)
GenoPath: A Pipeline to Infer Tumor Clone Composition, Mutational History, and Metastatic Cell Migration Events from Tumor DNA Sequencing Data
Frontiers in Bioinformatics | 5:1615834 (10 pp)
MEGA12: Molecular Evolutionary Genetics Analysis version 12 for Adaptive and Green Computing
Molecular Biology and Evolution | 41:msae263 (9 pp)
Discovering Fragile Clades And Causal Sequences In Phylogenomics By Evolutionary Sparse Learning
Molecular Biology and Evolution | 41:msae131 (13 pp)
Methods for Estimating Personal Disease Risk and Phylogenetic Diversity of Hematopoietic Stem Cells
Molecular Biology and Evolution | 41:msad279 (11 pp)
Visualizing Genomic Medicine: An Introduction to General Biology
The American Biology Teacher | 86:265–273 (9 pp)
Of Phylogenies and Tumors: Cancer as a Model System to Teach Evolution
The American Biology Teacher | 86:62–70 (9 pp)
Completing a Molecular Timetree of Primates
Frontiers in Bioinformatics | 4:1495417 (6 pp)
Some Mechanistic Underpinnings of Molecular Adaptations of SARS-COV-2 Spike Protein by Integrating Candidate Adaptive Polymorphisms with Protein Dynamics
eLife | 12:RP92063 (25 pp)
Analyses of Allele Age and Fitness Impact Reveal Human Beneficial Alleles to be Older than Neutral Controls
eLife | 13:RP93258 (27 pp)
Computational Reproducibility of Molecular Phylogenies
Molecular Biology and Evolution | 40:msad165 (9 pp)
Completing a Molecular Timetree of Apes and Monkeys
Frontiers in Bioinformatics | 3:1284744 (8 pp)
Molecular Timetrees using Relaxed Clocks and Uncertain Phylogenies
Frontiers in Bioinformatics | 3:1225807 (13 pp)
The Origin of Eukaryotes and Rise in Complexity were Synchronous with the Rise in Oxygen
Frontiers in Bioinformatics | 3:1233281 (10 pp)
Discovering Research Articles Containing Evolutionary Timetrees by Machine Learning
Bioinformatics | 39:btad035 (7 pp)
Improving Cellular Phylogenies through the Integrated Use of Mutation Order and Optimality Principles
Computational and Structural Biotechnology Journal | 21:3894–3903
Bootstrap Confidence for Molecular Evolutionary Estimates from Tumor Bulk Sequencing Data
Frontiers in Bioinformatics | 3:1090730 (11 pp)
Protein Dynamics Provide Mechanistic Insights about Epistasis among Common Missense Polymorphisms
Biophysical Journal | 122:2938–2947
Gene Expression Study of Breast Cancer using Welch Satterthwaite t-test, Kaplan-Meier Estimator Plot and Huber Loss Robust Regression Model
Journal of King Saud University - Science | 35:102447 (9 pp)
Masatoshi Nei (1931–2023)
Nature Ecology & Evolution | 7:(2 pp)
Masatoshi Nei (1931–2023)
Molecular Biology and Evolution | 40:msad149 (5 pp)
Embracing Green Computing in Molecular Phylogenetics
Molecular Biology and Evolution | 39:msac043 (4 pp)
TimeTree 5: An Expanded Resource for Species Divergence Times
Molecular Biology and Evolution | 39:msac174 (6 pp)
Taming the Selection of Optimal Substitution Models in Phylogenomics by Site Subsampling and Upsampling
Molecular Biology and Evolution | 39:msac236 (8 pp)
TopHap: Rapid Inference of Key Phylogenetic Structures from Common Haplotypes in Large Genome Collections with Limited Diversity
Bioinformatics | 38:2719–2726
Epistasis Creates Invariant Sites and Modulates the Rate of Molecular Evolution
Molecular Biology and Evolution | 39:msac106 (11 pp)
Dynamic Coupling of Residues within Proteins as a Mechanistic Foundation of Many Enigmatic Pathogenic Missense Variants
PLoS Computational Biology | 18:e1010006 (22 pp)
Limitations of Phylogenomic Data Can Drive Inferred Speciation Rate Shifts
Molecular Biology and Evolution | 39:msac038 (11 pp)
Storyboarding for Biology: An Authentic STEAM Experience
The American Biology Teacher | 84:328–335
Epistasis Storyboarded
The American Biology Teacher | 84:562–569
Clone Phylogenetics Reveals Metastatic Tumor Migrations, Maps, and Models
Cancers | 14:4326 (13 pp)
A Phylogenetic Approach to Study the Evolution of Somatic Mutational Processes in Cancer
Communications Biology | 5:617 (11 pp)
MOCA for Integrated Analysis of Gene Expression and Genetic Variation in Single Cells
Frontiers in Genetics | 13:831040 (8 pp)
Waiting for the Truth: Is Reluctance in Accepting an Early Origin Hypothesis for SARS-CoV-2 Delaying Our Understanding of Viral Emergence?
BMJ Global Health | 7:e008386 (8 pp)
Molecular Evidence for SARS-CoV-2 in Samples Collected from Patients with Morbilliform Eruptions since Late Summer 2019 in Lombardy, Northern Italy
Environmental Research | 215:113979 (9 pp)
Development of "Biosearch System" for Biobank Management and Storage of Disease Associated Genetic Information
Journal of King Saud University - Science | 34:101760 (10 pp)
An Evolutionary Portrait of the Progenitor SARS-CoV-2 and its Dominant Offshoots in COVID-19 Pandemic
Molecular Biology and Evolution | 38:3046–3059
Evolutionary Sparse Learning for Phylogenomics
Molecular Biology and Evolution | 38:4674–4682
Fast and Accurate Bootstrap Confidence Limits on Genome-scale Phylogenies using Little Bootstraps
Nature Computational Science | 1:573–577
Tumors are Evolutionary Island-like Ecosystems
Genome Biology and Evolution | 13:evab276 (11 pp)
Migrations of Cancer Cells through the Lens of Phylogenetic Biogeography
Scientific Reports | 11:17184 (13 pp)
Data-driven Speciation Tree Prior for Better Species Divergence Times in Calibration-poor Molecular Phylogenies
Bioinformatics | 37:i102–i110
Epistasis Produces an Excess of Invariant Sites in Neutral Molecular Evolution
Proceedings of the National Academy of Sciences (USA) | 118:e2018767118
MEGA11: Molecular Evolutionary Genetics Analysis Version 11
Molecular Biology and Evolution | 38:3022–3027
Molecular Dating for Phylogenies Containing a Mix of Populations and Species by Using Bayesian and RelTime Approaches
Molecular Ecology Resources | 21:122–136
How to Build a Super Predator: From Genotype to Phenotype
The American Biology Teacher | 83:138–146
Assessing Rapid Relaxed-Clock Methods for Phylogenomic Dating
Genome Biology and Evolution | 13:evab251 (14 pp)
Molecular and Morphological Clocks for Estimating Evolutionary Divergence Times
BMC Ecology and Evolution | 21:83 (15 pp)
Deep Low-Shot Learning for Biological Image Classification and Visualization from Limited Training Samples
IEEE Transactions on Neural Networks and Learning Systems | (11 pp)
Common Treatment, Common Variant: Evolutionary Prediction of Functional Pharmacogenomic Variants
Journal of Personalized Medicine | 11:131 (13 pp)
TreeMap: A Structured Approach to Fine Mapping of eQTL Variants
Bioinformatics | 37:1125–1134
The Durability of Immunity against Reinfection by SARS-CoV-2: A Comparative Evolutionary Study
The Lancet Microbe | 12:e666–e675
The Somatic Molecular Evolution of Cancer: Mutation, Selection, and Epistasis
Progress in Biophysics and Molecular Biology | 165:56–65
PathFinder: Bayesian Inference of Clone Migration Histories in Cancer
Bioinformatics | 36:i675–i683
Efficient Methods for Dating Evolutionary Divergences
in The Molecular Evolutionary Clock (ed. Ho) [Springer] | 197–219
Where Did SARS-CoV-2 Come From?
Molecular Biology and Evolution | 37:2463–2464
Molecular Evolutionary Genetics Analysis (MEGA) for macOS
Molecular Biology and Evolution | 37:1237–1239
A New Method for Inferring Timetrees from Temporally Sampled Molecular Sequences
PLoS Computational Biology | 16:e1007046 (24 pp)
Reliable Confidence Intervals for RelTime Estimates of Evolutionary Divergence Times
Molecular Biology and Evolution | 37:280–290
Power and Pitfalls of Computational Methods for Inferring Clone Phylogenies and Mutation Orders from Bulk Sequencing Data
Scientific Reports | 10:3498 (21 pp)
Relative Efficiencies of Simple and Complex Substitution Models in Estimating Divergence Times in Phylogenomics
Molecular Biology and Evolution | 37:1819–1831
Using a GTR+Gamma Substitution Model for Dating Sequence Divergence when Stationarity and Time-reversibility Assumptions are Violated
Bioinformatics | 36:i884–i894
The Bits and Bytes of Biology: Digitalization Fuels an Emerging Generative Platform for Biological Innovation
Handbook of Digital Innovation (ed. Nambisan, Lyytinen, Yoo) [Edward Elgar] | 253–265
Molecular Memories of a Cambrian Fossil
The American Biology Teacher | 82:586–595
Functional, Morphological, and Evolutionary Characterization of Hearing in Subterranean, Eusocial African Mole-Rats
Current Biology | 30:(18 pp)
Interactive Effect of TLR SNPs and Exposure to Sexually Transmitted Infections on Prostate Cancer Risk in Jamaican Men
The Prostate | 80:4329–4341.e4
Beaver and Naked Mole Rat Genomes Reveal Common Paths to Longevity
Cell Reports | 32:(13 pp)
Somatic Selection Distinguishes Oncogenes and Tumor Suppressor Genes
Bioinformatics | 36:1712–1717
The Role of Conformational Dynamics and Allostery in Modulating Protein Evolution
Annual Review of Biophysics | 49:267–288
Molecular Biology and Evolution of Cancer: From Discovery to Action
Molecular Biology and Evolution | 37:320–326
Deep Model Based Transfer and Multi-task Learning for Biological Image Analysis
IEEE Transactions on Big Data | 6:322–333
Biological Relevance of Computationally Predicted Pathogenicity of Noncoding Variants
Nature Communications | 10:330 (11 pp)
Delineation of Tumor Migration Paths by Using a Bayesian Biogeographic Approach
Cancers | 11:12 (14 pp)
Adventures in Evolution: The Narrative of Tardigrada, Trundlers in Time
The American Biology Teacher | 81:543–552
On Estimating Evolutionary Probabilities of Population Variants
BMC Evolutionary Biology | 19:133 (14 pp)
A Machine Learning Method for Detecting Autocorrelation of Evolutionary Rates in Large Phylogenies
Molecular Biology and Evolution | 36:811–824
Genome-wide Analysis Indicates Association between Heterozygote Advantage and Healthy Aging in Humans
BMC Genetics | 20:52 (14 pp)
MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms
Molecular Biology and Evolution | 35:1547–1549
Theoretical Foundation of the RelTime Method for Estimating Divergence Times from Variable Evolutionary Rates
Molecular Biology and Evolution | 35:1770–1782
Accurate Timetrees Require Accurate Calibrations
Proceedings of the National Academy of Sciences (USA) | 115:E9510–E9511
Neutral Theory, Disease Mutations, and Personal Exomes
Molecular Biology and Evolution | 35:1297–1303
Somatic Evolutionary Timings of Driver Mutations
BMC Cancer | 18:85 (10 pp)
RelTime Relaxes the Strict Molecular Clock throughout the Phylogeny
Genome Biology and Evolution | 10:1631–1636
Adaptive Landscape of Protein Variation in Human Exomes
Molecular Biology and Evolution | 35:2015–2025
Predicting Clone Genotypes from Tumor Bulk Sequencing of Multiple Samples
Bioinformatics | 34:4017–4026
Computational Enhancement of Single-cell Sequences for Inferring Tumor Evolution
Bioinformatics | 34:i917–i926
FlyExpress 7: An Integrated Discovery Platform to Study Coexpressed Genes Using in situ Hybridization Images in Drosophila
G3: Genes, Genomes, Genetics | 7:2791–2797
TimeTree: A Resource for Timelines, Timetrees, and Divergence Times
Molecular Biology and Evolution | 34:1812–1819
Fast and Accurate Estimates of Divergence Times from Big Data
Molecular Biology and Evolution | 34:45–50
The Reliability and Stability of an Inferred Phylogenetic Tree from Empirical Data
Molecular Biology and Evolution | 34:718–723
e-GRASP: An Integrated Evolutionary and GRASP Resource for Exploring Disease Associations
BMC Genomics | 17:770 (8 pp)
MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets
Molecular Biology and Evolution | 33:1870–1874
A Molecular Evolutionary Reference for the Human Variome
Molecular Biology and Evolution | 33:245–254
Advances in Time Estimation Methods for Molecular Data
Molecular Biology and Evolution | 33:863–869
Integration of Structural Dynamics and Molecular Evolution via Protein Interaction Networks: A New Era in Genomic Medicine
Current Opinion in Structural Biology | 35:135–142
Using Disease-associated Coding Sequence Variation to Investigate Functional Compensation by Human Paralogous Proteins
Evolutionary Bioinformatics | 11:245–251
A Protocol for Diagnosing the Effect of Calibration Priors on Posterior Time Estimates: A Case Study for the Cambrian Explosion of Animal Phyla
Molecular Biology and Evolution | 32:1907–1912
Conformational Dynamics of Nonsynonymous Variants at Protein Interfaces Reveals Disease Association
Proteins | 83:428–435
Phylogenetic Placement of Metagenomic Reads Using the Minimum Evolution Principle
BMC Genomics | 16:S13 (9 pp)
Evolutionary Diagnosis of Non-synonymous Variants Involved in Differential Drug Response
BMC Medical Genomics | 8:S6 (6 pp)
Tree of Life Reveals Clock-like Speciation and Diversification
Molecular Biology and Evolution | 32:835–845
Exceptional Reduction of the Plastid Genome of Saguaro Cactus (Carnegiea gigantea): Loss of the Ndh Gene Suite and Inverted Repeat
American Journal of Botany | 102:1115–1127
Prospects for Building Large Timetrees using Molecular Data with Incomplete Gene Coverage among Species
Molecular Biology and Evolution | 31:2542–2550
Signatures of Natural Selection on Mutations of Residues with Multiple Posttranslational Modifications
Molecular Biology and Evolution | 31:1641–1645
No Positive Selection for G Allele in a p53 Response Element in Europeans
Cell | 157:1497–1499
Reply to: "Proper Reporting of Predictor Performance"
Nature Methods | 11:781–782
myFX: A Turn-key Software for Laboratory Desktops to Analyze Spatial Patterns of Gene Expression in Drosophila Embryos
Bioinformatics | 30:1319–1321
MEGA-MD: Molecular Evolutionary Genetics Analysis Software with Mutational Diagnosis of Amino Acid Variation
Bioinformatics | 30:1305–1307
Automated Annotation of Developmental Stages of Drosophila Embryos in Images Containing Spatial Patterns of Expression
Bioinformatics | 30:266–273
Whole-genome Sequencing of the Snub-nosed Monkey Provides Insights into Folivory and Evolutionary History
Nature Genetics | 46:1303–1310
The Evolutionary History of Amino Acid Variations Mediating Increased Resistance of S. aureus Identifies Reversion Mutations in Metabolic Regulators
PLoS One | 8:e56466 (9 pp)
Structural Dynamics Flexibility Informs Function and Evolution at a Proteome Scale
Evolutionary Applications | 6:423–433
GRASP [Genomic Resource Access for Stoichioproteomics]: Comparative Explorations of the Atomic Content of 12 Drosophila Proteomes
BMC Genomics | 14:(14 pp)
Evolutionary Balancing is Critical for Correctly Forecasting Disease-associated Amino Acid Variants
Molecular Biology and Evolution | 30:1252–1257
Image-level and Group-level Models for Drosophila Gene Expression Pattern Annotation
BMC Bioinformatics | 14:350 (13 pp)
MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0
Molecular Biology and Evolution | 30:2725–2729
Genome-wide Profiling of Human Cap-independent Translation-enhancing Elements
Nature Methods | 10:747–750
A Mesh Generation and Machine Learning Framework for Drosophila Gene Expression Pattern Image Analysis
BMC Bioinformatics | 14:372 (10 pp)
Exploring Spatial Patterns of Gene Expression from Fruit Fly Embryogenesis on the iPhone
Bioinformatics | 28:2847–2848
Sharing and Re-use of Phylogenetic Trees (and associated data) to Facilitate Synthesis
BMC Research Notes | 5:574 (15 pp)
Evolutionary Meta-analysis of Association Studies Reveals Ancient Constraints Affecting Disease Marker Discovery
Molecular Biology and Evolution | 29:2087–2094
Human Genomic Disease Variants: A Neutral Evolutionary Explanation
Genome Research | 22:1383–1394
Performance of Computational Tools in Evaluating the Functional Impact of Laboratory-induced Amino Acid Mutations
Bioinformatics | 28:2093–2096
Comparison of Embryonic Expression within Multigene Families using the FlyExpress Discovery Platform Reveals More Spatial than Temporal Divergence
Developmental Dynamics | 241:150–160
Statistics and Truth in Phylogenomics
Molecular Biology and Evolution | 29:457–472
Evolutionary Diagnosis Method for Variants in Personal Exomes
Nature Methods | 9:855–856
MEGA-CC: Computing Core of Molecular Evolutionary Genetics Analysis Program for Automated and Iterative Data Analysis
Bioinformatics | 28:2685–2686
Drosophila Gene Expression Pattern Annotation through Multi-instance Multi-label Learning
IEEE/ACM Transactions on Computational Biology and Bioinformatics | 9:98–112
Purifying Selection Modulates the Estimates of Population Differentiation and Confounds Genome-wide Comparisons across Single-nucleotide Polymorphisms
Molecular Biology and Evolution | 29:3617–3623
Estimating Divergence Times in Large Molecular Phylogenies
Proceedings of the National Academy of Sciences (USA) | 109:19333–19338
Learning Sparse Representations for Fruit-fly Gene Expression Pattern Image Annotation and Retrieval
BMC Bioinformatics | 13:107 (15 pp)
Molecular Clock: Testing
eLS | (7 pp)
Fast and Slow Implementations of Relaxed-clock Methods Show Similar Patterns of Accuracy in Estimating Divergence Times
Molecular Biology and Evolution | 28:2439–2442
Stoichiogenomics: The Evolutionary Ecology of Macromolecular Elemental Composition
Trends in Ecology & Evolution | 26:38–44
Rampant Purifying Selection Conserves Positions with Posttranslational Modifications in Human Proteins
Molecular Biology and Evolution | 28:1565–1568
Phylomedicine: An Evolutionary Telescope to Explore and Diagnose the Universe of Disease Mutations
Trends in Genetics | 27:377–386
FlyExpress: Visual Mining of Spatiotemporal Patterns for Genes and Publications in Drosophila Embryogenesis
Bioinformatics | 27:3319–3320
Evolution of Modern Birds Revealed by Mitogenomics: Timing the Radiation and Origin of Major Orders
Molecular Biology and Evolution | 28:1927–1942
MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods
Molecular Biology and Evolution | 28:2731–2739
Performance of Relaxed-clock Methods in Estimating Evolutionary Divergence Times and Their Credibility Intervals
Molecular Biology and Evolution | 27:1289–1300
Quantitative Analysis of the Drosophila Segmentation Regulatory Network Using Pattern Generating Potentials
PLoS Biology | 8:(19 pp)
More Reliable Estimates of Divergence Times in Pan Using Complete mtDNA Sequences and Accounting for Population Structure
Philosophical Transactions of the Royal Society B: Biological Sciences | 365:3277–3288
Ecological Nitrogen Limitation Shapes the DNA Composition of Plant Genomes
Molecular Biology and Evolution | 26:953–956
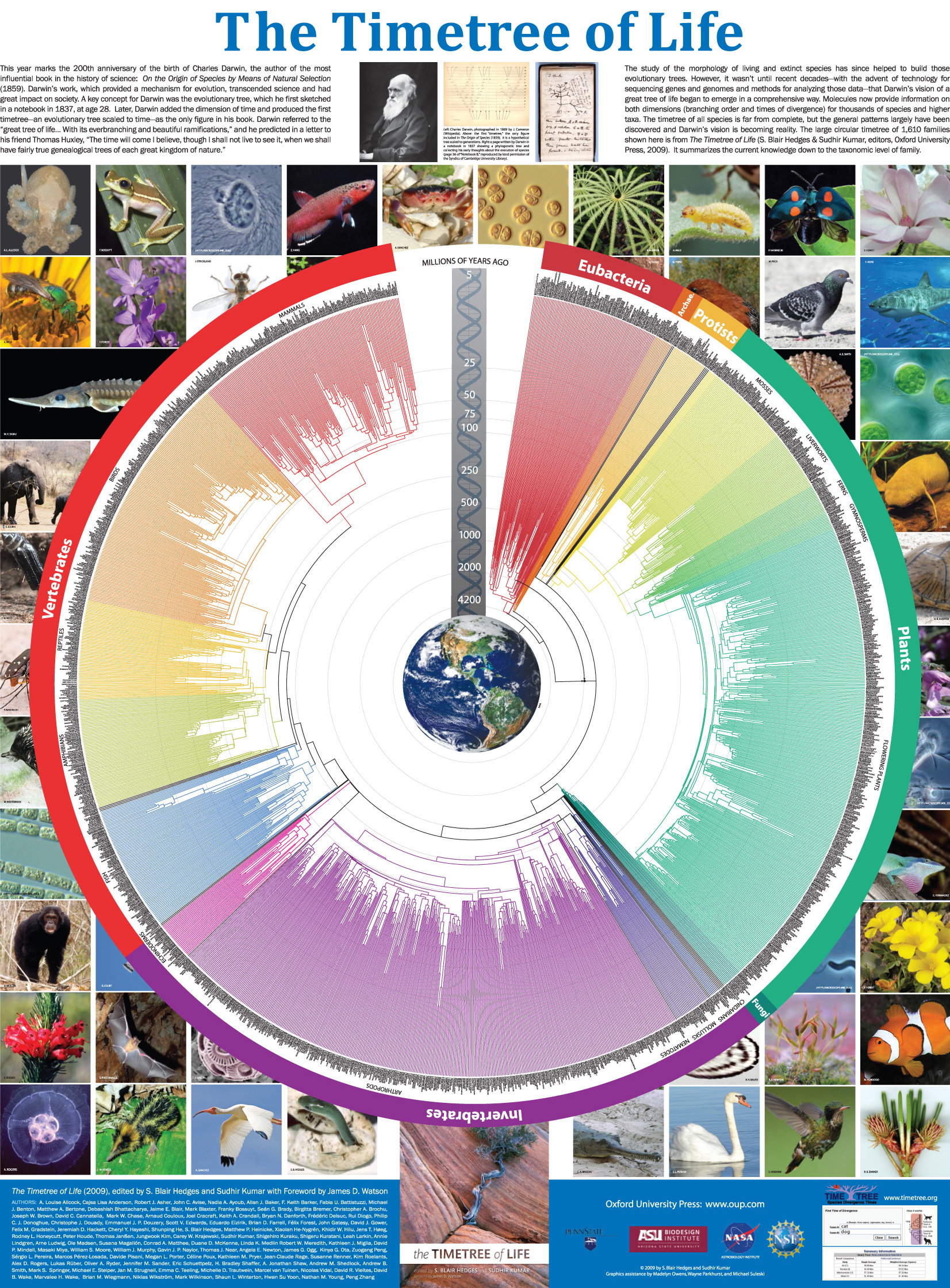
The Timetree of Life
Oxford University Press (New York) | (551 pp)


Discovering the Timetree of Life
The TimeTree of Life (ed. Hedges, Kumar) [Oxford University Press] | 3–18
Signatures of Nitrogen Limitation in the Elemental Composition of the Proteins Involved in the Metabolic Apparatus
Proceedings of the Royal Society B: Biological Sciences | 276:2605–2610
Association of Orthodenticle with Natural Variation for Early Embryonic Patterning in Drosophila melanogaster
Journal of Experimental Zoology Part B: Molecular and Developmental Evolution | 312:841–854
Phylogenetic Construction of 17 Bacterial Phyla by New Method and Carefully Selected Orthologs
Gene | 429:59–64
A Bag-of-words Approach for Drosophila Gene Expression Pattern Annotation
BMC Bioinformatics | 10:119 (16 pp)
Drosophila Gene Expression Pattern Annotation Using Sparse Features and Term-term Interactions
KDD-09: 15th ACM SIGKDD Conference on Knowledge Discovery and Data Mining | 407–415
Positional Conservation and Amino Acids Shape the Correct Diagnosis and Population Frequencies of Benign and Damaging Personal Amino Acid Mutations
Genome Research | 19:1562–1569
Relationship between Gene Co-expression and Sharing of Transcription Factor Binding Sites in Drosophila Melanogasterc
Bioinformatics | 25:2473–2477
The Origin of Metazoa: A Transition from Temporal to Spatial Cell Differentiation
BioEssays | 31:758–768
Methods for Incorporating the Hypermutability of CpG Dinucleotides in Detecting Natural Selection Operating at the Amino Acid Sequence Level
Molecular Biology and Evolution | 26:2275–2284
Developmental Stage Annotation of Drosophila Gene Expression Pattern Images via an Entire Solution Path for LDA
ACM Transactions on Knowledge Discovery from Data | 2:(21 pp)
Biological Image Analysis via Matrix Approximation
Encyclopedia of Data Warehousing and Mining (ed. Wang) [Idea Group] | 166–170
Automated Annotation of Drosophila Gene Expression Patterns Using a Controlled Vocabulary
Bioinformatics | 24:1881–1888
MEGA: A Biologist-centric Software for Evolutionary Analysis of DNA and Protein Sequences
Briefings in Bioinformatics | 9:299–306
Detecting Molecular Signatures of Adaptive Evolution
Evolutionary Genomics and Proteomics (ed. Pagel, Pomiankowski) [Sinauer] | 241–254
Nullomers: Really a Matter of Natural Selection?
PLoS One | 2:1022–1025
Evolution of Genes and Genomes on the Drosophila Phylogeny
Nature | 450:203–218
Lower Bounds on Multiple Sequence Alignment Using Exact 3-way Alignment
BMC Bioinformatics | 8:140 (8 pp)
Bioinformatics Software for Biologists in the Genomics Era
Bioinformatics | 23:1713–1717
Multiple Sequence Alignment: In Pursuit of Homologous DNA Positions
Genome Research | 17:127–135
MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) Software Version 4.0
Molecular Biology and Evolution | 24:1596–1599
Constraint and Turnover in Sex-biased Gene Expression in the Genus Drosophila
Nature | 450:233–237
Classification of Drosophila Embryonic Developmental State Range Based on Gene Expression Pattern Images
Computational Systems Bioinformatics Conference | 2006:293–298
Codon-based Detection of Positive Selection Can be Biased by Heterogeneous Distribution of Polar Amino Acids along Protein Sequences
Computational Systems Bioinformatics Conference | 2006:335–340
Signatures of Ecological Resource Availability in the Animal and Plant Proteomes
Molecular Biology and Evolution | 23:1946–1951
TimeTree: A Public Knowledge-base of Divergence Times among Organisms
Bioinformatics | 22:2971–2972
Constraining Fossil Calibrations for Molecular Clocks
BioEssays | 28:770–771
Evolutionary Anatomies of Positions and Types of Disease-associated and Neutral Amino Acid Mutations in the Human Genome
BMC Genomics | 7:306 (9 pp)
Higher Intensity of Purifying Selection on >90% of the Human Genes Revealed by the Intrinsic Replacement Mutation Rates
Molecular Biology and Evolution | 23:2283–2287
The Evolution of the Genome: Comparative Genomics in Eukaryotes
The Evolution of the Genome (ed. Gregory) [Elsevier] | 521–583
Maximum Likelihood Outperforms Maximum Parsimony Even When Evolutionary Rates are Heterotachous
Molecular Biology and Evolution | 22:2139–2141
Inferring Species Phylogenies from Multiple Genes: Concatenated Sequence Tree versus Consensus Gene Tree
Journal of Experimental Zoology Part B: Molecular and Developmental Evolution | 304:64–74
Automatic Annotation Techniques for Gene Expression Images of the Fruit Fly Embryo
Visual Communications and Image Processing 2005, Pts 1-4 | 5960:576–583
Molecular Clocks: Four Decades of Evolution
Nature Reviews Genetics | 6:654–662
Placing Confidence Limits on the Molecular Age of the Human-Chimpanzee Divergence
Proceedings of the National Academy of Sciences (USA) | 102:18842–18847
Pushing Back the Expansion of Introns in Animal Genomes
Cell | 123:1182–1184
Image Registration and Similarity Computation for Chicken Gene Expression Patterns
Genomic Signal Processing and Statistics | (4 pp)
The Spectrum of Human Rhodopsin Disease Mutations through the Lens of Interspecific Variation
Gene | 12:107–118
Identifying Spatially Similar Gene Expression Patterns in Early Stage Fruit Fly Embryo Images: Binary Feature versus Invariant Moment Digital Representations
BMC Bioinformatics | 16:202–214
Precision of Molecular Time Estimates
Trends in Genetics | 20:242–247
Gene Expression Intensity Shapes Evolutionary Rates of the Proteins Encoded by the Vertebrate Genome
Genetics | 168:373–381
Prospects for Inferring Very Large Phylogenies by Using the Neighbor-joining Method
Proceedings of the National Academy of Sciences (USA) | 101:11030–11035
Temporal Patterns of Fruit Fly (Drosophila) Evolution Revealed by Mutation Clocks
Molecular Biology and Evolution | 21:36–44
MEGA3: Integrated Software for Molecular Evolutionary Genetics Analysis and Sequence Alignment
Briefings in Bioinformatics | 5:150–163
Genomic Clocks and Evolutionary Timescales
Trends in Genetics | 19:200–206
Genomic Sequence of a Ranavirus (family Iridoviridae) Associated with Salamander Mortalities in North America
Virology | 316:90–103
Quantifying the Intragenic Distribution of Human Disease Mutations
Annals of Human Genetics | 67:567–579
Heterogeneity of Nucleotide Frequencies among Evolutionary Lineages and Phylogenetic Inference
Molecular Biology and Evolution | 20:610–621
Taxon Sampling, Bioinformatics, and Phylogenomics
Systematic Biology | 52:119–124
Patterns of Transitional Mutation Biases within and among Mammalian Genomes
Molecular Biology and Evolution | 20:988–993
Neutral Substitutions Occur at a Faster Rate in Exons than in Noncoding DNA in Primate Genomes
Genome Research | 13:838–844
Vertebrate Genomes Compared
Science | 297:1283–1285
Measuring Conservation of Contiguous Sets of Autosomal Markers on Bovine and Porcine Genomes in Relation to the Map of the Human Genome
Genome | 45:769–776
BEST: A Novel Computational Approach for Comparing Gene Expression Patterns from Early Stages of Drosophila melanogaster Development
Genetics | 162:2037–2047
Mutation Rates in Mammalian Genomes
Proceedings of the National Academy of Sciences (USA) | 99:803–808
Evolutionary Distance Estimation under Heterogeneous Substitution Pattern among Lineages
Molecular Biology and Evolution | 19:1727–1736
Elucidating Gene Interaction Networks Based on Gene Expression Pattern Image Analysis
Proceedings of the International Conference on Biomedical Engineering | 5:232–234
Does Nonneutral Evolution Shape Observed Patterns of DNA Variation in Animal Mitochondrial Genomes?
Annual Review of Genetics | 35:539–566
A Genomic Timescale for the Origin of Eukaryotes
BMC Evolutionary Biology | 1:4 (10 pp)
Mutation and Linkage Disequilibrium in Human mtDNA
European Journal of Human Genetics | 9:969–972
Classification and Indexing of Gene Expression Images
Applications of Digital Image Processing XXIV | 4472:471–481
Disparity Index: A Simple Statistic to Measure and Test the Homogeneity of Substitution Patterns between Molecular Sequences
Genetics | 158:1321–1327
Determination of the Number of Conserved Chromosomal Segments between Species
Genetics | 157:1387–1395
MEGA2: Molecular Evolutionary Genetics Analysis Software
Bioinformatics | 17:1244–1245
Understanding Human Disease Mutations through the Use of Interspecific Genetic Variation
Human Molecular Genetics | 10:2319–2328
Incomplete Taxon Sampling is Not a Problem for Phylogenetic Inference
Proceedings of the National Academy of Sciences (USA) | 98:10751–10756
Traditional Phylogenetic Reconstruction Methods Reconstruct Shallow and Deep Evolutionary Relationships Equally Well
Molecular Biology and Evolution | 18:1823–1827
Efficiency of the Neighbor-joining Method in Reconstructing Deep and Shallow Evolutionary Relationships in Large Phylogenies
Journal of Molecular Evolution | 51:544–553
Questioning Evidence for Recombination in Human Mitochondrial DNA
Science | 288:1931a (2 pp)
Expansion and Molecular Evolution of the Interferon-induced 2 '-5 ' Oligoadenylate Synthetase Gene Family
Molecular Biology and Evolution | 17:738–750
Single Column Discrepancy and Dynamic Max-Mini Optimizations for Quickly Finding the Most Parsimonious Evolutionary Trees
Bioinformatics | 16:140–151
Molecular Evolution of a Developmental Pathway: Phylogenetic Analyses of Transforming Growth Factor-beta Family Ligands, Receptors and Smad Signal Transducers
Genetics | 152:783–795
Divergence Time Estimates for the Early History of Animal Phyla and the Origin of Plants, Animals and Fungi
Proceedings of the Royal Society B: Biological Sciences | 266:163–171
Divergence Times of Eutherian Mammals
Science | 285:2031a (2 pp)
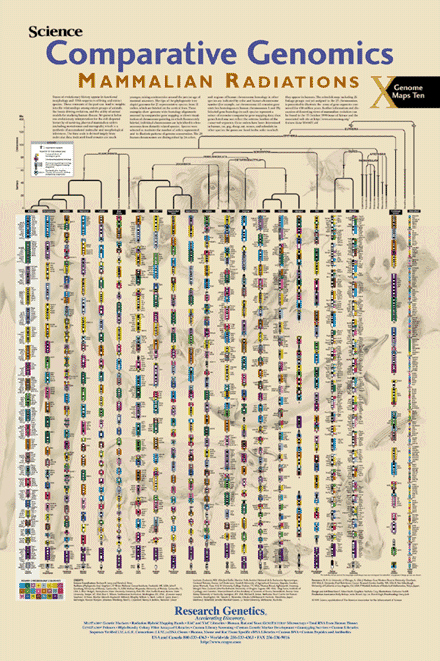
Genome Maps 10. Comparative Genomics. Mammalian Radiations. Wall Chart.
Science | 286:463–478

A Molecular Timescale for Vertebrate Evolution
Nature | 392:917–920
The Optimization Principle in Phylogenetic Analysis Tends to Give Incorrect Topologies When the Number of Nucleotides or Amino Acids Used is Small
Proceedings of the National Academy of Sciences (USA) | 95:12390–12397
Evolution and Functional Diversification of the Paired Box (Pax) DNA-binding Domains
Molecular Biology and Evolution | 14:829–842
Detection of Convergent and Parallel Evolution at the Amino Acid Sequence Level
Molecular Biology and Evolution | 14:527–536
Sequence Convergence in the Peptide-binding Region of Primate and Rodent MBC Class Ib Molecules
Molecular Biology and Evolution | 14:1035–1041
Tempo and Mode of Nucleotide Substitutions in GAG and ENV Gene Fragments in Human Immunodeficiency Virus Type 1 Populations with a Known Transmission History
Journal of Virology | 71:4761–4770
Small-sample Tests of Episodic Adaptive Evolution: A Case Study of Primate Lysozymes
Molecular Biology and Evolution | 14:1335–1338
Stepwise Algorithm for Finding Minimum Evolution Trees
Molecular Biology and Evolution | 13:584–593
Patterns of Nucleotide Substitution in Mitochondrial Protein Coding Genes of Vertebrates
Genetics | 143:537–548
Evolutionary Relationships of Eukaryotic Kingdoms
Journal of Molecular Evolution | 42:183–193
Evolution of the Hedgehog Gene Family
Genetics | 142:965–972
Approximate Methods for Estimating the Pattern of Nucleotide Substitution and the Variation of Substitution Rates among Sites
Molecular Biology and Evolution | 13:650–659
Continental Breakup and the Ordinal Diversification of Birds and Mammals
Nature | 381:226–229
A New Method of Inference of Ancestral Nucleotide and Amino Acid Sequences
Genetics | 141:1641–1650
Four-cluster Analysis: A Simple Method to Test Phylogenetic Hypotheses
Molecular Biology and Evolution | 12:163–167
18S Ribosomal-RNA Data Indicate that Aschelminthes are Polyphyletic in Origin and Consist of at Least Three Distinct Clades
Molecular Biology and Evolution | 12:1132–1137
MEGA - Molecular Evolutionary Genetics Analysis Software for Microcomputers
Computer Applications in the Biosciences | 10:189–191
A Guide to Molecular Evolutionary Genetics Analysis Program for Microcomputers
Pennsylvania State University [University Park, PA, USA] | (135 pp)
Human Origins and Analysis of Mitochondrial DNA Sequences
Science | 255:737–739